 Model Architecture
Model ArchitectureAbstract
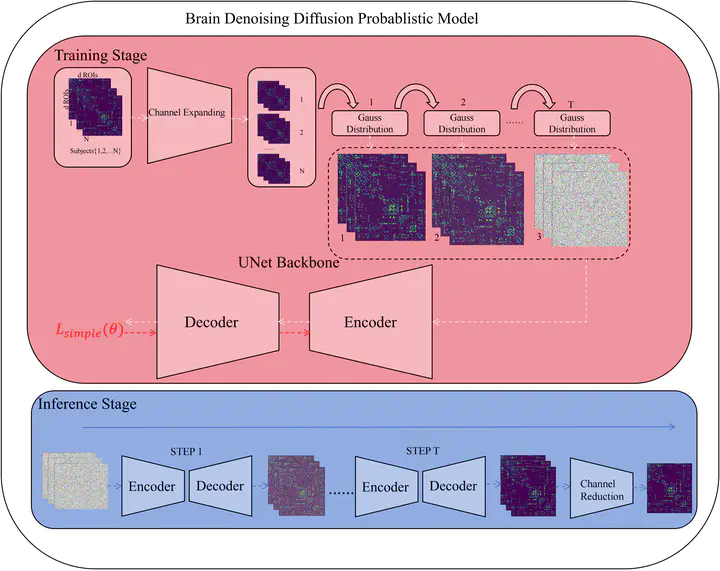
Alzheimer’s disease (AD) significantly impairs the quality of life for a vast patient population and poses treatment challenges, partly due to its elusive pathophysiological mechanisms. The analysis of structural brain networks is fundamental to elucidating these mechanisms. However, acquiring data on such networks is non-trivial, hindered by the slow generation of structural brain network data and the complexities involved in obtaining DTI, a key requisite for their construction. In this study, we introduce a brain denoising diffusion probabilistic model designed to synthesize structural brain networks at various stages of AD, thereby mitigating the difficulties inherent in data acquisition. We trained this model on the ADNI dataset, utilizing two sets of data: one comprising structural brain networks produced by PANDA, and the other amalgamating these PANDA-generated networks with those synthesized by our model. Both datasets were employed to train a DiffPool for subsequent diagnostic tasks. It can be seen that the brain networks generated by the brain denoising diffusion probabilistic model are beneficial for structural brain networks in downstream diagnostic tasks.