 Model Architecture
Model ArchitectureAbstract
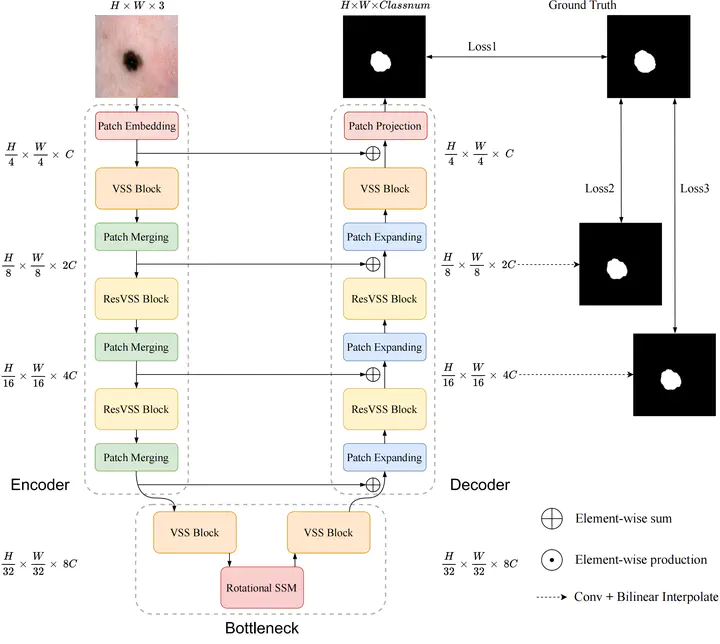
Accurate segmentation of tissues and lesions is crucial for disease diagnosis, treatment planning, and surgical navigation. Yet, the complexity of medical images presents significant challenges for traditional Convolutional Neural Networks (CNNs) and Transformer models due to their limited receptive fields or high computational complexity. State Space Models (SSMs) have recently shown notable vision performance, particularly Mamba and its variants. However, their feature extraction methods may not be sufficiently effective and retain some redundant structures, leaving room for parameter reduction. In response to these challenges, we introduce a methodology called Rotational Mamba-UNet, characterized by Residual Visual State Space (ResVSS) block and Rotational SSM Module. The ResVSS block is devised to mitigate network degradation caused by the diminishing efficacy of information transfer from shallower to deeper layers. Meanwhile, the Rotational SSM Module is devised to tackle the challenges associated with channel feature extraction within State Space Models. Finally, we propose a weighted multi-level loss function, which fully leverages the outputs of the decoder’s three stages for supervision. We conducted experiments on ISIC17, ISIC18, CVC-300, Kvasir-SEG, CVC-ColonDB, Kvasir-Instrument datasets, and Low-grade Squamous Intraepithelial Lesion (LSIL) datasets provided by The Third Affiliated Hospital of Sun Yat-sen University, demonstrating the superior segmentation performance of our proposed RM-UNet. Additionally, compared to the previous VM-UNet, our model achieves a one-third reduction in parameters.